How Communication Among Cells Affects Development of Multicellular Tissue
Oct 15, 2018 — Atlanta, GA
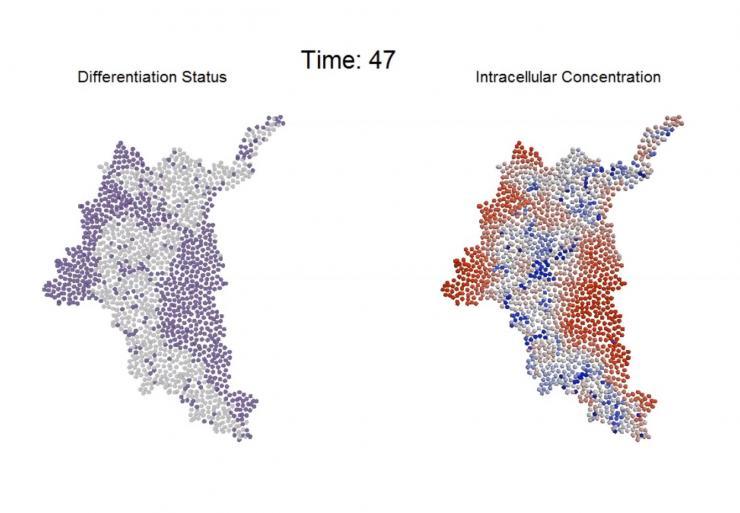
An agent-based computational model simulates the progression of intracellular molecular gradients and resultant differentiation patterns that emerge within a multicellular system. (Credit: Chad Glen, Georgia Tech)
Using a combination of computational modeling and experimental techniques, a research team has developed new information about how intercellular communication affects the differentiation of an embryonic stem cell colony over time.
By providing new information about the role of communication among cells, the research could lead to a better understanding of how multicellular organoids form from colonies of independent cells. The information could lead to new methods for controlling how multicellular constructs develop, and that could have applications in regenerative medicine, pharmaceutical testing and other research areas.
The research resulted from collaboration between the Georgia Institute of Technology and the Gladstone Institutes, and was reported October 5 in the journal Nature Communications. The National Science Foundation’s (NSF) Emergent Behaviors of Integrated Cellular Systems Science and Technology Center (EBICS) supported the research.
“The goal is to control a system of cells like this to direct tissues to take on different phenotypes, to develop into different complex mixtures, and to self-assemble and emerge into very complicated structures,” said Melissa Kemp, an associate professor in the Wallace H. Coulter Department of Biomedical Engineering at Georgia Tech and Emory University. “For developing tissues that could be used as surrogates for screening drugs or as eventual implants for therapeutic purposes, we need to how to control and direct them properly.”
Scientists believe that the patterning of stem cell differentiation affects what kinds of cells will ultimately emerge from the differentiation process.
Despite the importance of local cell-to-cell interactions in evolving multicellular systems, little has been known about how the overall system regulates its morphological processes. To learn more about this, the paper’s first author, Chad Glen, studied how communication between adjacent mouse cells informs the fate of those cells. Beyond understanding this communication, Glen discovered a potential mechanism for “braking” the rate of differentiation without affecting the overall patterning of the resulting multicellular tissue.
“The amount of coordination among cells that are tightly coupled gives us an idea about how they work as a group,” said Todd McDevitt, senior investigator at the Gladstone Institutes and a professor of bioengineering and therapeutic sciences at the University of California, San Francisco. “This reflects the behavior of a team versus individuals. They really do coordinate activity in a rapid way. This study shows how quickly some important cell behaviors are mediated by gap junction communication.”
Glen, a recent Ph.D. graduate of Georgia Tech, began the project by studying communication between pairs of adjacent cells, which have pores that allow small molecules to enter. By introducing a signaling molecule into a colony containing hundreds of these homogeneous mouse stem cells, the researchers observed that differentiation began with a change in a single cell. That cell triggered a pattern of differentiation that flowed through the cells and eventually led to changes in the entire colony.
On a larger scale and in three dimensions, such changes lead to development of bodily organs. Understanding how that happens – and how it could be controlled – could be key to directing this transition from individual cells.
Based on experimental observations, Glen developed a model of the process, which allowed the researchers to study the impact of a series of interrelated variables that would have been impossible to study experimentally. The modeling of several hundred individual cells led to specific predictions that the researchers then tested experimentally.
To understand and measure the cell communication process, the researchers used a fluorescent dye to show when signaling molecules had moved from one cell to its neighbors. They then used a laser to bleach the dye from a single cell. Measuring how long it took for the dye to be replaced showed the permeability of the cell membrane – and how well the cell was communicating with its neighbors.
“If you zap a cell and it immediately returns to green, you know there is a lot of fluidity and cross-talk between the cell membranes,” Kemp explained. “If you zap a cell and it stays dark, the cell is effectively isolated and has no communication with its neighbors.”
By partially blocking communication between cells and otherwise perturbing the communications, the researchers slowed the differentiation process – but didn’t change its pattern. “We were able to shift the way the cells were behaving to slow things down – effectively ‘braking’ the process – while preserving the spatial information,” said Kemp. “Cells that were at 48 hours in the process could look like 24-hour cells.”
The combination of computational modeling and experiment allowed the research team to arrive at answers that neither technique by itself could have provided, McDevitt noted.
“With modeling, you can study a much larger set of conditions and parameters than we could experimentally,” he said. “The model could make predictions that we could go back and study experimentally to see how those conditions actually affected the behaviors. The behaviors we measured experimentally matched what the computer predicted, and validated that we had a robust model.”
McDevitt and Kemp have been studying this system of embryonic stem cells for several years, and the new study moves them closer to a more complete understanding of the complex system.
“We have demonstrated in a series of papers over the past six years that this system’s complexity can be modeled,” he said. “This paper represents a further step toward the greater goal of integrating information about this system. With each of these steps, we are much closer to taking a bigger leap into potentially controlling these systems.”
This research was supported by the National Science Foundation Emergent Behaviors of Integrated Cellular Systems Science and Technology Center (CBET 0939511). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation.
CITATION: Chad M. Glen, Todd C. McDevitt, Melissa L. Kemp, “Dynamic intercellular transport modulates the spatial patterning of differentiation during early neural commitment, (Nature Communications 9, 2018). http://dx.doi.org/10.1038/s41467-018-06693-1
Research News
Georgia Institute of Technology
177 North Avenue
Atlanta, Georgia 30332-0181 USA
Media Relations Contact: John Toon (404-894-6986) (jtoon@gatech.edu).
Writer: John Toon
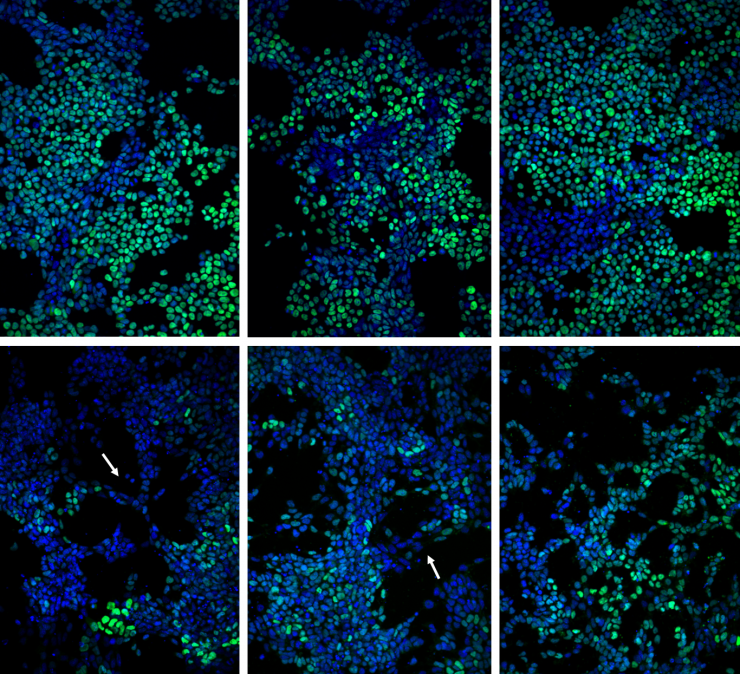
Image shows colony patterning and morphology after 24 and 48 hours of retinoic acid exposure, respectively, with adenylyl cyclase or gap junction inhibition compared to the vehicle control. (Credit: Chad Glen, Georgia Tech)
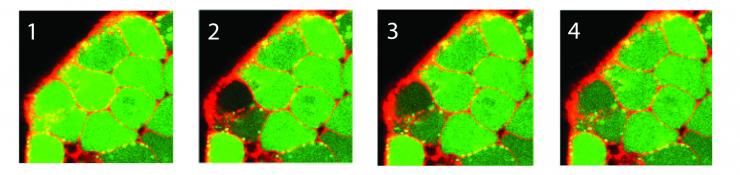
A sequence of images depicts the photo bleaching (frame 2) and recovery (frames 3-4) of a fluorescent dye for quantification of relative diffusion rates between adjacent stem cells within a colony. (Credit: Chad Glen, Georgia Tech)
John Toon
Research News
(404) 894-6986




